DNA microarray, also called DNA chips, gene chips, DNA arrays, gene arrays and biochips, are microscopic slides of glass or silicon printed with thousands of small spots in grid fashion with each containing known DNA or gene. Each slide acts as probe to detect gene expression. Basically, it’s been evolved from southern blotting. It is different from Southern blotting as here the probe is fixed/attached, and sample DNA is labeled rather than probe.
PRINCIPLE OF DNA MICROARRAY
The basic principle behind DNA microarray lies on Nucleic acid hybridization. During this method, two complementary strands of DNA are joined together by hydrogen bond to make a double stranded molecule by hydrogen bond. Restriction endonuclease is employed to cut the unknown DNA molecules into small fragments. Fluorescent markers are attached to the fragments, and these get react with probes in DNA chips. DNA probes are then binds with the target DNA with complementary sequences and unbounded DNA fragments are washed away. Identification of the target pieces of DNA is done by their fluorescence emission passing through a laser beam and computer recorded the pattern of emission as well as DNA identification.
TYPES OF DNA MICROARRAY
There are four types of DNA microarray:
- Oligo DNA microarray: It uses oligonucleotides of 20-50 nucleotides long. Oligonucleotides are synthesized directly on the slide. Single color hybridization used for each probe. It has good specifcity but poor sensitivity.
- cDNA microarray: It is usually referred to as spotted microarray in within which DNA fragments of any length (500bp-1kb) or oligos of 20-100 nts are stuck to the glassslides. It uses two colors hybridization for every probe.
- BAC Microarray: It uses the template which is amplified by polymerase chain reaction as the probe.
- SNA Microarray: It is used to detect polymorphisms within a population.
REQUIREMENTS:
- 1. DNA Chip
- 2. Target sample
- 3. Sample
- 4. Enzymes
- 5. Fluorescent dyes
- 6. Probes
PREPARATION OF MICROARRAY
There are several methods which are used to prepare a spotted array. Readymade probes are attached to the fine needles and are printed on a chemical matrix surface using a robot (surface engineering). Other methods are photoactivated chemistry and masking and these can also be purchased from commercially.
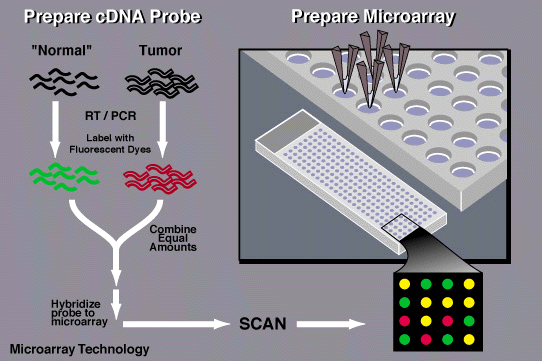
PROCEDURE OF MICROARRAY BASED ON cDNA.
1. Collection of sample
The sample may be a cell/tissue of the organisms. Two types of samples are collected; one healthy cells and another infected cells.
2. Isolation of mRNA
RNA is extracted from sample using a column or solvent like phenol-chloroform. Then mRNA is separated from the extracted RNA leaving behind rRNA and tRNA. Column beads with poly-T tails are used to bind mRNA as mRNA has a poly –A tail. During extraction, the column is rinsed with buffer to isolate mRNA from the beads.
3. Creation of labeled cDNA
A labeling is added to RNA that contains the mixture of poly-T (oligo dT) , primers, reverse transcriptase (t make cDNA), and fluorescently dyed nucleotides. Cyanine 3(Cy3-fluoresces green) is added to the healthy cells and cyanine 5 (cy5-fluoresces red) to infected cells. Firstly, the primer and RT bind to the mRNA and then fluorescently dyed nucleotides are added which creates a complementary strand of DNA.
- Hybridization
The labeled cDNA from both the samples are placed in the DNA microarray so that each cDNA gets hybridized to its complementary strand and also thoroughly washed to remove unbounded sequences.
- Collection and analysis
Data is collected by using a microarray scanner which consist of a laser, a computer and a camera. The laser excites fluorescence of cDNA and recording is done by camera while the laser scans the array and finally data is stored by computer and gives result immediately. The given data is analyzed and the difference in the intensity of the color for each spots determines the character of the gene in that particular spot.
*Green-the healthy sample hybridize more than the diseased sample
*Red-the diseased sample hybridized more than the non diseased
*Yellow-Both samples hybridized equally to the target DNA
Applications of DNA Microarray
- Gene expression and profiling: It is employed in detection of the presence and absence of specific genes, compare genes from different sources and to know how the external genes affect the external stimuli. It is used to detect the gene expression pattern of different diseases such as cancer. It is also helpful in studying the effect of certain treatments, diseases and developmental stage on gene expression.
- Disease Diagnosis: It helps the researchers to investigate about different diseases such as heart diseases, infectious diseases, mental illness and cancer. Due to the advancement in microarray technology, it makes very easy to know the types of cancer on the basis of gene acting in tumor cells.
- Gene Discovery: This technology is used in identification of new genes and their functions as well as their expression level in various conditions.
- Drug Discovery: It is extensively applied in Pharmacogenomics. The comparative analysis of genes help to identify specific proteins which is produced by diseased cell resulting to synthesize drug that combat with these proteins and help to reduce their effects.
- Gene ID: It is used to check IDs of food organisms and also to feed Mycoplasmas in cell culture usually by combining microarray and PCR technology.
- Research: It is beneficial in conducting different researches related to toxicogenomic studies and nutrigenmic studies. It is also used to reveal the presence of antibiotic resistance genes and to study microbial pathogenicity.
LIMITATIONS of DNA Microarray
- The initial high cost for the synthesis of gene specific primers, fluorescence used in labeling is the major limitation.
- The shelf life of the chips -are not very long.
- The analysis of results become complex as too many results at a time.
Learn more about
REFERENCES
- Melissa B., Milter and Yi-Wei Tang. Basic Concepts of Microarrays and Potential Applications in Clinical Microbiology, Oct.2009, p.611-633
- Dari Shalon, Stephen J. Smirth, and Patrick O.Brown, A DNA Microarray system for Analyzing Complex DNA Samples Using Two-color Fluoresent Probe Hybridization,Genome Res. 6:639-645,2017
- Lena . Stru B, Ulla Ruffing, Salim Abdulla , et.al 2016, Detecting Staphylococus aureus Virulence and Resistance Genes. Journal of Clinical Microbiology Vol.54 Issue no. 4, pg 1008-1016.S
- Schena, M., Shalon, D., Davis, R., & Brown, P. (1995). Quantitative Monitoring of Gene Expression Patterns with a Complementary DNA Microarray. Science, 270(5235),467-470. doi: 10.1126/science.270.5235.467
- Drachici, S., Khatri, P., Eklund, A., & Szallasi, Z. (2006). Reliability and reproducibility issues in DNA microarray measurements. Trends In Genetics, 22(2), 101-109. doi: 10.1016/j.tig.2005.12.005
- Bustin, S.A. (ed.). A-Z of Quantitative PCR. (International University Line Biotechnology Series, La Jolla, California, USA, 2004).
- Wilhelm, M. et al. Array-based comparative genomic hybridization for the differential diagnosis of renal cell cancer. Cancer Res. 62, 957–960 (2002).